Aug
10
Algae Metabolism Is Now Documented
August 10, 2011 | Leave a Comment
In a collaboration of universities across the U.S., Iceland and the United Arab Emirates the metabolic network of the algae strain Chlamydomonas reinhardtii has been documented. This will provide a new platform for genomic data analysis and phenotype prediction.
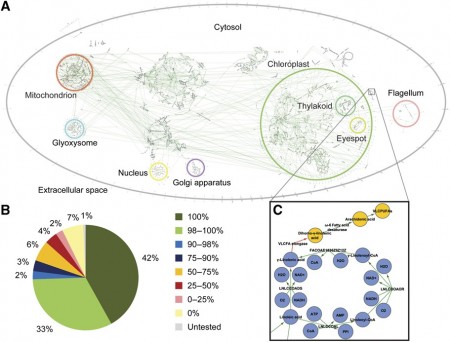
A metabolic network reconstruction encompasses existing knowledge about an organism’s metabolism and the genome.
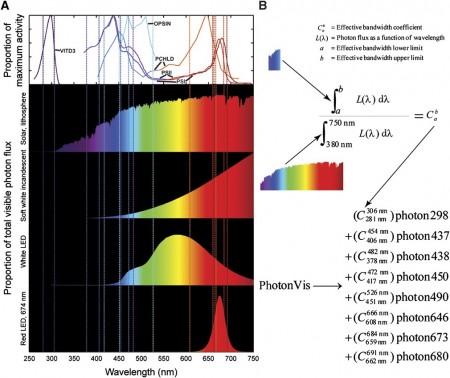
The research offers some concrete results. By integrating biological and optical data, the team reconstructed a genome-scale metabolic ‘network’ for the alga and devised a novel light-modeling approach that enabled quantitative growth prediction for a given light source, revealing the best wavelength and photon flux. The team experimentally checked basic information assumptions in the network and physiologically validated model function through simulation and generation of new experimental growth data.
The experimental checks provide high confidence in ‘network’ contents and predictive applications. The network offers insight into algal metabolism and potential for genetic engineering and efficient light source design, a pioneering resource for studying light-driven metabolism and quantitative systems biology.
Commercial scale production and further basic scientific research of light energized bio growth should benefit from better understanding of how light is absorbed and how it affects cell’s photosynthesis. The quality of light sources arriving to the organisms in commercial production photobioreactors largely determines the efficiency of energy usage in industrial algal farming. Light spectral quality also affects how photon absorption induces various metabolic processes: photosynthesis, pigment and vitamin synthesis, and the retinol pathway required for phototaxis.
Using a metabolic network reconstruction can provide a framework to integrate a wide range of input data for investigation of most of the properties of metabolism. This ability can provide clear advantages in ways to study the effects of light upon a photosynthetic biological system when light input is accounted for by itself.
This first research experimental check revealed hypothetical latent pathways of lipid metabolism, the functions of which have likely been lost through evolution. The first experiments also found the suggestion that C. reinhardtii also lacks very long-chain fatty acids (VLCFAs), their polyunsaturated analogs (VLCPUFAs) and ceramides. While these haven’t been detected in C. reinhardtii, the absence is now confirmed by another route.
The paper goes into considerable detail outlining the experiment and the evidence obtained on the metabolism of C. reinhardtii. What the research offers is an insight on the means by which C. reinhardtiiis operates metabolically, both from its evolutionary past and into the potential engineered future.
The most impressive work is learning what are the effective light spectral ranges by analyzing biochemical activity spectra, either reaction activity or absorbance at varying light wavelengths. Defining effective spectral bandwidths associated with each photon-utilizing reaction enabled the network to model growth under different light sources. Now the possibility exists to distinguish the amount of emitted photons that drive different metabolic reactions. This can open the door to optimally using solar light, various light bulbs, and LEDs.
The research is quite broad and inclusive, including the thermal aspect and sets an impressive standard for further research into the models of particular organisms. Where the shine comes is the breadth and depth of the experiments allow for a future platform for prediction of phenotypic outcomes of system perturbations, light source evaluation and design, and genetic engineering design for production of biofuels and other commodity chemicals.
With a base set now, which will surely find further research and more insight for C. reinhardtiiis, this alga has a early start in the genetic engineering arena. The complete genome sequence data for C. reinhardtii and its functional annotation have enabled the research team’s effort to build a more full understanding of the presence of genome-encoded enzymes. The team has put together and experimentally validated a genome-scale metabolic network, the first research to account for detailed photon absorption permitting growth simulations under different light sources. This surprise is the new research shows the activity of substantially more genes with metabolic functions than previous research.
This is just the first run as well. Exciting as it is, this research is on a single cell organism. Where this caliber of research might lead on the more complex plants, of particular interest food and other fuel plants producing fruit, seeds and other products, is a whole new frontier.